Research
My research results and code snippets.
Table of contents
Technical Research Poster
Presented this poster to share my research findings at the American Geophysical Union (AGU) 2021 conference. I utilized Adobe InDesign to create the poster and python to create the figures.
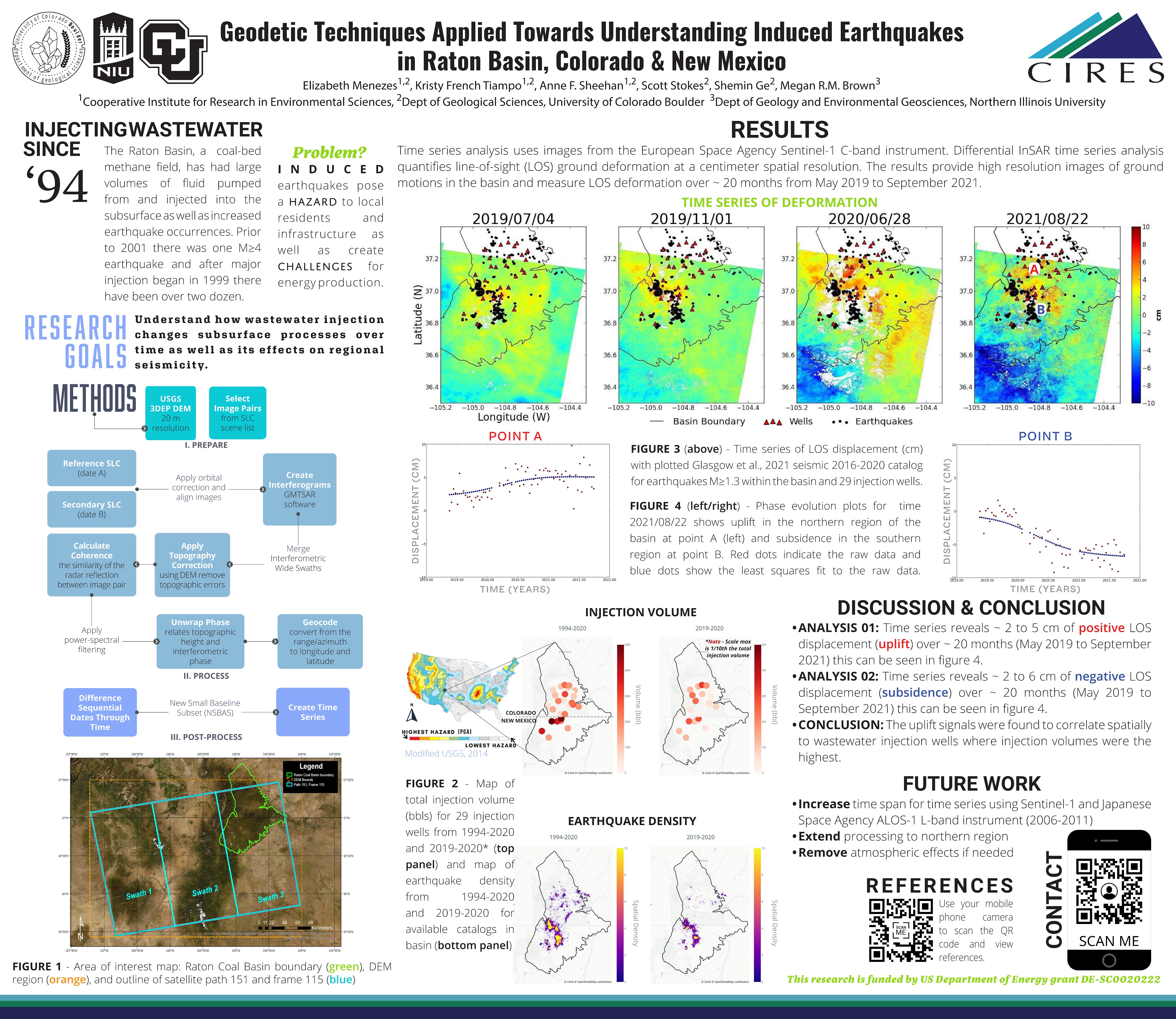
Wastewater Injection Well Density Maps
In python, I made these cumulative density maps to easily show the locations of the highest injection volumes in the Raton Basin.
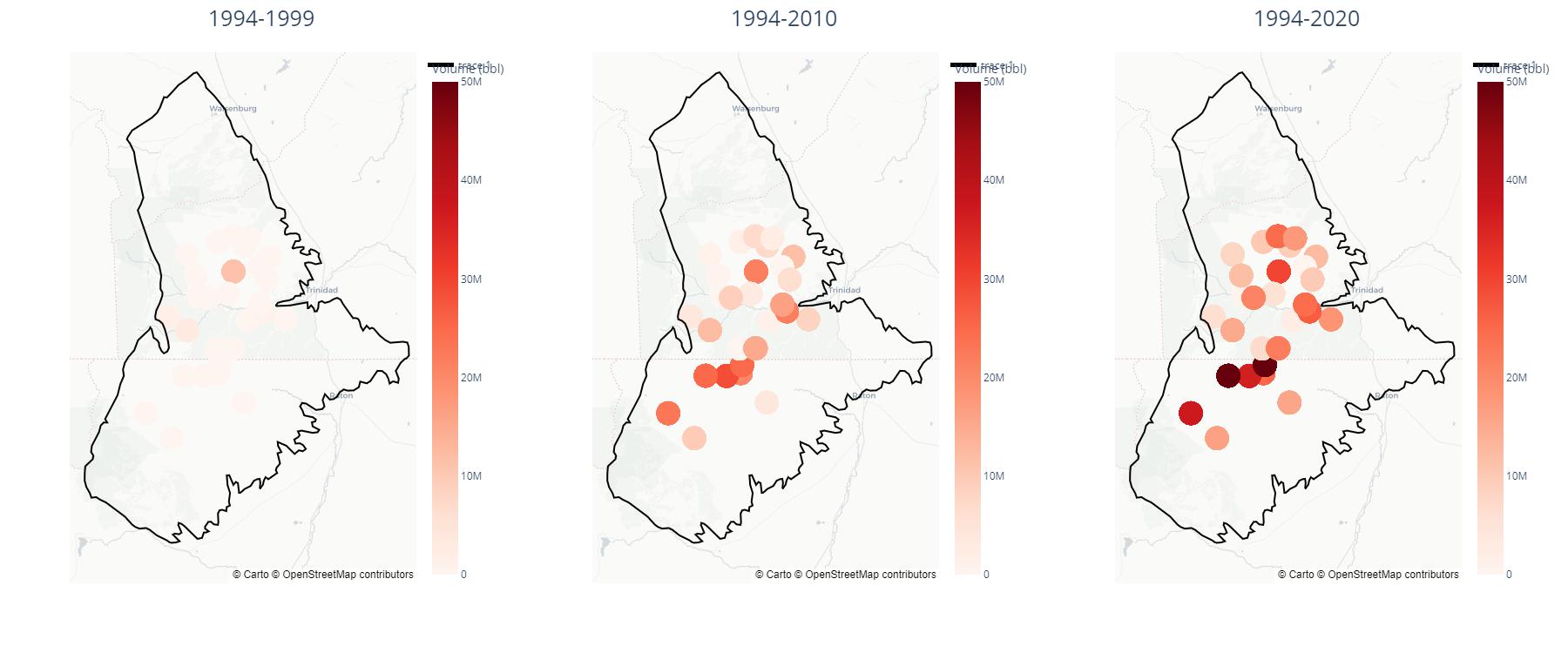 —
—
Data for 29 Injection Wells
Retrived data from the following sources for wastewater injection wells located in the Raton basin and concatenated the data in to a table called ‘CatInjection.csv’.
Colorado Oil and Gas Conservation Commission (COGCC) [Retrieved 2020]: Facility > County code: 071 > Sequence Code: “second # in API#” > Submit > Well > click Name > Export production to Excel > water in bbl/mo
New Mexico Oil Conservation Division (OCD)(NMOCD) [Retrieved 2020]: Enter API Number: 07 - “second # in API#” > Continue > scroll to bottom > Export to Excel
Code Snippet
My code is shown here:
## Cumulative Injection Volume Density Map
# Import libraries
import geopandas as gpd
import pandas as pd
import plotly.express as px
import plotly.graph_objects as go
import plotly.subplots as sp
import os
import numpy as np
import PIL
from PIL import Image
# Plot density map 1
df = pd.read_csv('CatInjection.csv', header= 0,
encoding= 'unicode_escape')
size = 10
max_size =20
# Read data period 1994-1999
start_date = 1994
end_date = 1999
mask = (df['year'] >= start_date) & (df['year'] <= end_date)
df = df.loc[mask]
def f(x):
x['Volume (bbl)'] = (x['vol']).sum()
return x
df_new = df.groupby(['name']).apply(f)
size_for_plot = [size]*len(df_new)
df_new['size_for_plot'] = size_for_plot
figure1 = px.scatter_mapbox(df_new, lat='lat', lon='lon', color="Volume (bbl)", size='size_for_plot',
color_continuous_scale=px.colors.sequential.Reds, size_max=max_size,
center=dict(lat=37.1, lon=-104.75), zoom=8, range_color=[0, 50000000],
mapbox_style="carto-positron")
# pd.set_option('display.max_rows', df.shape[0]+1)
# df_new
# Plot boundary
bound = go.Figure(go.Scattermapbox(
mode="lines",
fill="none",
lon=lon_all,
lat=lat_all,
hoverinfo='none',
marker={'size': 10, 'color': "black"}))
figure1.add_traces(bound.data)
figure1.update_layout(title="1994-1999", title_font_size=25, title_x=0.5, title_xanchor="center",
width=600,
height=750)
# Plot density map 2
df = pd.read_csv('CatInjection.csv', header= 0,
encoding= 'unicode_escape')
# Read data period 1994-2010
start_date = 1994
end_date = 2010
mask = (df['year'] >= start_date) & (df['year'] <= end_date)
df = df.loc[mask]
def f(x):
x['Volume (bbl)'] = (x['vol']).sum()
return x
df_new = df.groupby(['name']).apply(f)
size_for_plot = [size]*len(df_new)
df_new['size_for_plot'] = size_for_plot
figure2 = px.scatter_mapbox(df_new, lat='lat', lon='lon', color="Volume (bbl)", size='size_for_plot',
color_continuous_scale=px.colors.sequential.Reds, size_max=max_size,
center=dict(lat=37.1, lon=-104.75), zoom=8, range_color=[0, 50000000],
mapbox_style="carto-positron")
# Plot boundary
bound = go.Figure(go.Scattermapbox(
mode="lines",
fill="none",
lon=lon_all,
lat=lat_all,
hoverinfo='none',
marker={'size': 10, 'color': "black"}))
figure2.add_traces(bound.data)
figure2.update_layout(title="1994-2010", title_font_size=25, title_x=0.5, title_xanchor="center",
width=600,
height=750)
# Plot density map 3
df = pd.read_csv('CatInjection.csv', header= 0,
encoding= 'unicode_escape')
# Read data period 1994-2020
start_date = 1994
end_date = 2020
mask = (df['year'] >= start_date) & (df['year'] <= end_date)
df = df.loc[mask]
average = df['year']
def f(x):
x['Volume (bbl)'] = (x['vol']).sum()
return x
df_new = df.groupby(['name']).apply(f)
size_for_plot = [size]*len(df_new)
df_new['size_for_plot'] = size_for_plot
figure3 = px.scatter_mapbox(df_new, lat='lat', lon='lon', color="Volume (bbl)", size='size_for_plot',
color_continuous_scale=px.colors.sequential.Reds, size_max=max_size,
center=dict(lat=37.1, lon=-104.75), zoom=8, range_color=[0, 50000000],
mapbox_style="carto-positron")
# Plot boundary
bound = go.Figure(go.Scattermapbox(
mode="lines",
fill="none",
lon=lon_all,
lat=lat_all,
hoverinfo='none',
marker={'size': 10, 'color': "black"}))
figure3.add_traces(bound.data)
figure3.update_layout(title="1994-2020", title_font_size=25, title_x=0.5, title_xanchor="center",
width=600,
height=750)
# show figure
figure1.show()
figure2.show()
figure3.show()
# create dir
if not os.path.exists("wellDensityImagesCum"):
os.mkdir("wellDensityImagesCum")
# save PNG
figure1.write_image("wellDensityImagesCum/fig1.png")
figure2.write_image("wellDensityImagesCum/fig2.png")
figure3.write_image("wellDensityImagesCum/fig3.png")
# align images
list_im = ['wellDensityImagesCum/fig1.png', 'wellDensityImagesCum/fig2.png', 'wellDensityImagesCum/fig3.png']
images = [ PIL.Image.open(i) for i in list_im ]
widths, heights = zip(*(i.size for i in images))
total_width = sum(widths)
max_height = max(heights)
new_im = Image.new('RGB', (total_width, max_height))
x_offset = 0
for im in images:
new_im.paste(im, (x_offset,0))
x_offset += im.size[0]
# save that beautiful picture
new_im.save('wellDensityImagesCum/Wellfigs.jpg')
References:
3D Inversion Fault Model
Data for Earthquake Catalog
Retrieved from the supplementary material in Nakai et al. 2017.
Code Snippet
In MATLAB, I made a 3D diagram of the earthquakes in my research field site. For my method in grouping earthquakes spatially together, I used a clustering algorithm in MATLAB to index the different fault lineations. My code is shown here:
%% Setup the Import Options and import the data
opts = delimitedTextImportOptions("NumVariables", 15);
% Specify range and delimiter
opts.DataLines = [2, Inf];
opts.Delimiter = ",";
% Specify column names and types
opts.VariableNames = ["year", "month", "day", "hour", "minute", "second", "lat", "lon", "depth", "north_sd", "east_sd", "depth_sd", "time_sd", "ML", "rms"];
opts.VariableTypes = ["double", "double", "double", "double", "double", "double", "double", "double", "double", "double", "double", "double", "double", "double", "double"];
% Specify file level properties
opts.ExtraColumnsRule = "ignore";
opts.EmptyLineRule = "read";
% Import the data
D = readtable("...\raton_nakai_eqs.csv", opts);
% Clear temporary variables
clear opts
% Import Raton Basin Boundary
B=shaperead('rbbndg.shp','usegeo', true);
% mapshow(B)
% convert degrees to km
latb = B.Lat';
latb(370,:) = [];
lonb = B.Lon';
lonb(370,:) = [];
db = zeros(1,369)';
% assign data variables
lat=D(:,7); % deg
lon=D(:,8); % deg
depth=D(:,9); % km
north_sd=D(:,10); % deg
east_sd=D(:,11); % deg
depth_sd=D(:,12); % km
% Standard deviations of north, east, depth and time are described
% as well as origin parameters, magnitude, and rms.
% Location method as described in this paper (Bayesloc) with velocity models as given in Table 2.
% Event depth is given relative to local surface elevation.
% table2array
lat=table2array(lat); % deg
lon=table2array(lon); % deg
depth=table2array(depth); % km
north_sd=table2array(north_sd); % km
east_sd=table2array(east_sd); % km
depth_sd=table2array(depth_sd); % km
% convert degrees to km
x = 111.12*cos((pi/180)*mean(lat))*(lon-min(lon));
y = 111.12*(lat-min(lat));
xb = 111.12*cos((pi/180)*mean(lat))*(lonb-min(lon));
yb = 111.12*(latb-min(lat));
dobs=-depth;
N=length(-depth);
%% 3D plot of data
% use rotate button to examine it prom different perspectives!
figure(1);
clf;
set(gca,'LineWidth',3);
set(gca,'FontSize',14);
hold on;
pxmin=-25; pxmax=75;
pymin=-25; pymax=110;
pzmin=-30; pzmax=0;
axis( [pxmin, pxmax, pymin, pymax, pzmin, pzmax]' );
plot3(x,y,dobs,'ko','LineWidth',1);
% Draw boundary
hold on;
plot3(xb,yb,db,'k','LineWidth',2,'DisplayName','Basin Boundary');
%% Group event clusters
idx = clusterdata([x,y,dobs],10);
gu=unique(idx);
clrstr={'#0aff99','#ff8700','#ffff3f','#147df5','r','r','r','r','#be0aff','r'};
clr=clrstr(1,1:length(gu));
mrksle = {'.','.','.','.','x','x','x','x','.','x'};
mrksze = 20;
mrkfclr = 'auto';
leg = false;
legloc = 'SouthEast';
hold on;
gscatter3(x,y,dobs,idx,clr,mrksle, mrksze,mrkfclr, leg, legloc)
hold on;
%% Draw 3D box
hold on;
plot3( [pxmin,pxmin], [pymin,pymin], [pzmin,pzmax], 'k-', 'LineWidth', 2 );
plot3( [pxmin,pxmin], [pymin,pymax], [pzmin,pzmin], 'k-', 'LineWidth', 2 );
plot3( [pxmin,pxmax], [pymin,pymin], [pzmin,pzmin], 'k-', 'LineWidth', 2 );
plot3( [pxmax,pxmax], [pymax,pymax], [pzmax,pzmin], 'k-', 'LineWidth', 2 );
plot3( [pxmax,pxmax], [pymax,pymin], [pzmax,pzmax], 'k-', 'LineWidth', 2 );
plot3( [pxmax,pxmin], [pymax,pymax], [pzmax,pzmax], 'k-', 'LineWidth', 2 );
plot3( [pxmax,pxmin], [pymin,pymin], [pzmax,pzmax], 'k-', 'LineWidth', 2 );
plot3( [pxmax,pxmax], [pymin,pymin], [pzmax,pzmin], 'k-', 'LineWidth', 2 );
plot3( [pxmin,pxmin], [pymax,pymin], [pzmax,pzmax], 'k-', 'LineWidth', 2 );
plot3( [pxmin,pxmin], [pymax,pymax], [pzmax,pzmin], 'k-', 'LineWidth', 2 );
plot3( [pxmax,pxmax], [pymax,pymin], [pzmin,pzmin], 'k-', 'LineWidth', 2 );
plot3( [pxmax,pxmin], [pymax,pymax], [pzmin,pzmin], 'k-', 'LineWidth', 2 );
%% LEGEND
legend('events','Basin Boundary', 'Cluster 1', 'Cluster 2','Cluster 3','Cluster 4','Cluster 5','Cluster 6','Cluster 7','Cluster 8','Cluster 9','Cluster 10');
% title('Fault Lineation Model')
xlabel('Latitude [km]')
ylabel('Longitude [km]')
zlabel('Depth [km]')
hold off;
%% RECORD
daspect([1,1,.3]);axis tight;
OptionZ.FrameRate=15;OptionZ.Duration=8;OptionZ.Periodic=false;
CaptureFigVid([0,90;0,90;-20,80;-40,60;-80,40;-100,20;-120,10;-140,10;-160,10;-180,10;...
-200,10;-220,10;-240,10;-260,10;-280,10;-300,10;-320,10;-340,10;-360,10],'Results(1)_test',OptionZ)